Output from Rgenetics QC report tool run at 19/05/2010 15:15:46
(Click any preview image to download a full sized PDF version)
Click here to download the Marker QC Detail report file (1.4 KB) tab delimited Click here to download the Subject QC Detail report file (1.4 KB) tab delimited 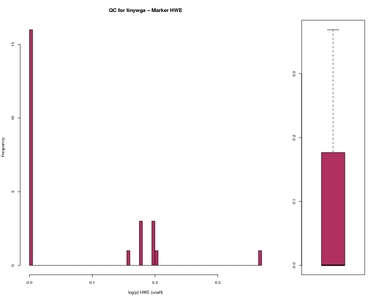
All tinywga QC Plots joined into a single pdf 
All tinywga QC Plots 3 by 3 to a page 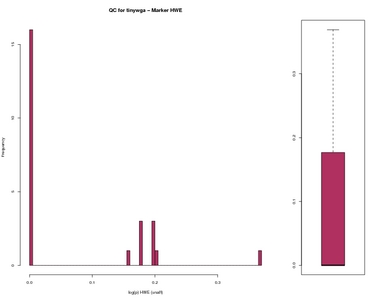
Marker HWE Worst data 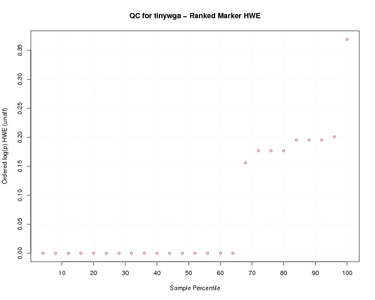
Ranked Marker HWE 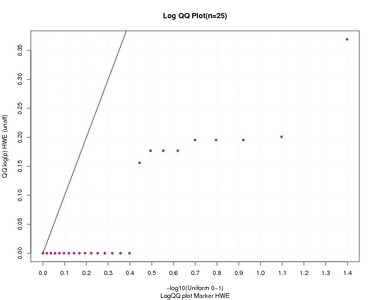
LogQQ plot Marker HWE 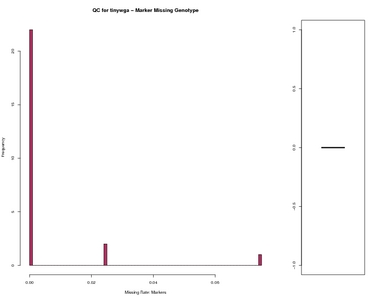
Marker Missing Genotype Worst data 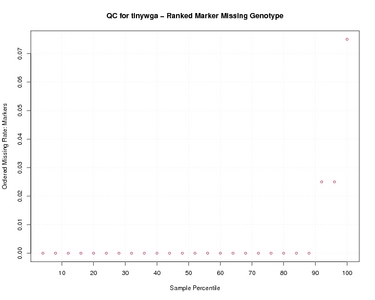
Ranked Marker Missing Genotype 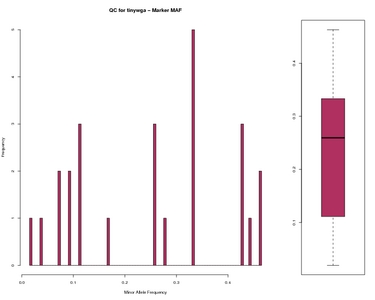
Marker MAF Worst data 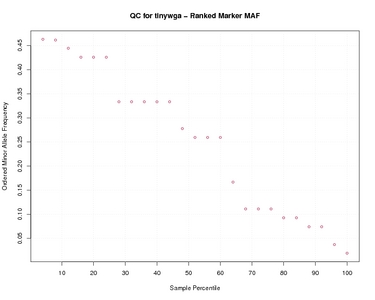
Ranked Marker MAF 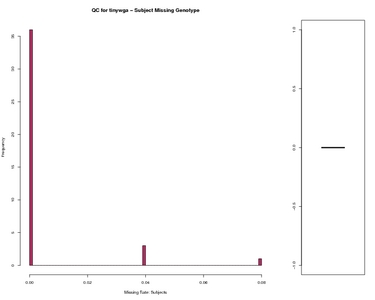
Subject Missing Genotype Worst data 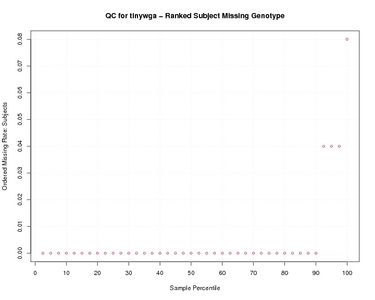
Ranked Subject Missing Genotype 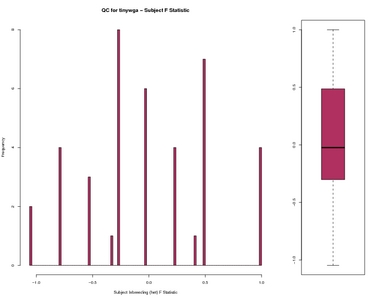
Subject F Statistic 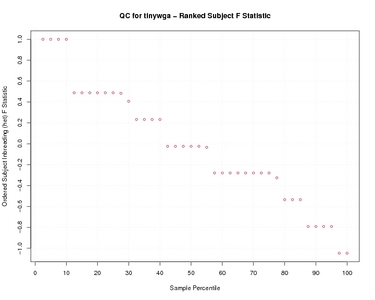
Ranked Subject F Statistic 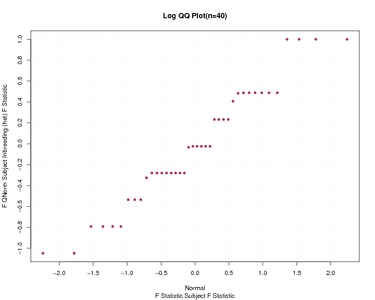
F Statistic Subject F Statistic
All output files from the QC run are available below
QC run log contents
## subject reports starting at 19/05/2010 15:15:38
## imissfile /opt/galaxy/test-data/rgtestouts/rgQC/tinywga.imiss contained 40 ids
### writing /opt/galaxy/test-data/rgtestouts/rgQC/SubjectDetails_rgQCtest1.xls report with ['famId', 'iId', 'FracMiss', 'Mendel_errors', 'Ped_sex', 'SNP_sex', 'Status', 'XHomEst', 'F_Stat']## marker reports starting at 19/05/2010 15:15:38
hwe header testpos=2,ppos=8,snppos=1
## starting plotpage, newfpath=/opt/galaxy/test-data/rgtestouts/rgQC,m=[['snp', 'chromosome', 'offset', 'maf', 'a1', 'a2', 'missfrac', 'p_hwe_all', 'logp_hwe_all', 'p_hwe_unaff', 'logp_hwe_unaff', 'N_Mendel'], ['rs2283802', '22', '21784722', '0.2593', '4', '2', '0', '0.638', '0.195179', '0.638', '0.195179', '0']],s=[['famId', 'iId', 'FracMiss', 'Mendel_errors', 'Ped_sex', 'SNP_sex', 'Status', 'XHomEst', 'F_Stat'], ['101', '1', '0.04', '0', '2', '0', 'PROBLEM', 'nan', '-0.03355']]/n## Rgenetics: http://rgenetics.org Galaxy Tools rgQC.py Plink runner
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--freq
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Writing allele frequencies (founders-only) to [ tinywga.frq ]
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--hwe 0.0
--missing
--hardy
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Writing Hardy-Weinberg tests (founders-only) to [ tinywga.hwe ]
0 markers to be excluded based on HWE test ( p <= 0 )
0 markers failed HWE test in cases
0 markers failed HWE test in controls
Writing individual missingness information to [ tinywga.imiss ]
Writing locus missingness information to [ tinywga.lmiss ]
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--mendel
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
14 nuclear families, 1 founder singletons found
13 non-founders with 2 parents in 13 nuclear families
0 non-founders without 2 parents in 0 nuclear families
10 affected offspring trios
0 phenotypically discordant parent pairs found
Converting data to Individual-major format
Writing all Mendel errors to [ tinywga.mendel ]
Writing per-offspring Mendel summary to [ tinywga.imendel ]
Writing per-family Mendel summary to [ tinywga.fmendel ]
Writing per-locus Mendel summary to [ tinywga.lmendel ]
0 Mendel errors detected in total
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--check-sex
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Converting data to Individual-major format
Writing X-chromosome sex check results to [ tinywga.sexcheck ]
Analysis finished: Wed May 19 15:15:38 2010
## Rgenetics: http://rgenetics.org Galaxy Tools rgQC.py Plink pruneLD runner
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile /opt/galaxy/test-data/tinywga
--indep-pairwise 40 20 0.5
--out tinywga
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Performing LD-based pruning...
Writing pruned-in SNPs to [ tinywga.prune.in ]
Writing pruned-out SNPs to [ tinywga.prune.out ]
Scanning from chromosome 22 to 22
Scan region on chromosome 22 from [ rs2283802 ] to [ rs4822375 ]
For chromosome 22, 11 SNPs pruned out, 14 remaining
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ ldp_tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile /opt/galaxy/test-data/tinywga
--extract tinywga.prune.in
--make-bed
--out ldp_tinywga
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Reading list of SNPs to extract [ tinywga.prune.in ] ... 14 read
Before frequency and genotyping pruning, there are 14 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.991071
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 14 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Writing pedigree information to [ ldp_tinywga.fam ]
Writing map (extended format) information to [ ldp_tinywga.bim ]
Writing genotype bitfile to [ ldp_tinywga.bed ]
Using (default) SNP-major mode
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile ldp_tinywga
--het
--out tinywga
Reading map (extended format) from [ ldp_tinywga.bim ]
14 markers to be included from [ ldp_tinywga.bim ]
Reading pedigree information from [ ldp_tinywga.fam ]
40 individuals read from [ ldp_tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ ldp_tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 14 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.991071
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 14 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
**Warning** this analysis typically requires whole-genome level data
to give accurate results
Converting data to Individual-major format
Writing individual heterozygosity information to [ tinywga.het ]
Analysis finished: Wed May 19 15:15:38 2010
(Click any preview image to download a full sized PDF version)
| Click here to download the Marker QC Detail report file (1.4 KB) tab delimited | ||
| Click here to download the Subject QC Detail report file (1.4 KB) tab delimited | ||

| All tinywga QC Plots joined into a single pdf | |

| All tinywga QC Plots 3 by 3 to a page | |

| Marker HWE | Worst data |

| Ranked Marker HWE | |

| LogQQ plot Marker HWE | |

| Marker Missing Genotype | Worst data |

| Ranked Marker Missing Genotype | |

| Marker MAF | Worst data |

| Ranked Marker MAF | |

| Subject Missing Genotype | Worst data |

| Ranked Subject Missing Genotype | |

| Subject F Statistic | |

| Ranked Subject F Statistic | |

| F Statistic Subject F Statistic | |
All output files from the QC run are available below
QC run log contents
## subject reports starting at 19/05/2010 15:15:38
## imissfile /opt/galaxy/test-data/rgtestouts/rgQC/tinywga.imiss contained 40 ids
### writing /opt/galaxy/test-data/rgtestouts/rgQC/SubjectDetails_rgQCtest1.xls report with ['famId', 'iId', 'FracMiss', 'Mendel_errors', 'Ped_sex', 'SNP_sex', 'Status', 'XHomEst', 'F_Stat']## marker reports starting at 19/05/2010 15:15:38
hwe header testpos=2,ppos=8,snppos=1
## starting plotpage, newfpath=/opt/galaxy/test-data/rgtestouts/rgQC,m=[['snp', 'chromosome', 'offset', 'maf', 'a1', 'a2', 'missfrac', 'p_hwe_all', 'logp_hwe_all', 'p_hwe_unaff', 'logp_hwe_unaff', 'N_Mendel'], ['rs2283802', '22', '21784722', '0.2593', '4', '2', '0', '0.638', '0.195179', '0.638', '0.195179', '0']],s=[['famId', 'iId', 'FracMiss', 'Mendel_errors', 'Ped_sex', 'SNP_sex', 'Status', 'XHomEst', 'F_Stat'], ['101', '1', '0.04', '0', '2', '0', 'PROBLEM', 'nan', '-0.03355']]/n## Rgenetics: http://rgenetics.org Galaxy Tools rgQC.py Plink runner
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--freq
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Writing allele frequencies (founders-only) to [ tinywga.frq ]
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--hwe 0.0
--missing
--hardy
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Writing Hardy-Weinberg tests (founders-only) to [ tinywga.hwe ]
0 markers to be excluded based on HWE test ( p <= 0 )
0 markers failed HWE test in cases
0 markers failed HWE test in controls
Writing individual missingness information to [ tinywga.imiss ]
Writing locus missingness information to [ tinywga.lmiss ]
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--mendel
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
14 nuclear families, 1 founder singletons found
13 non-founders with 2 parents in 13 nuclear families
0 non-founders without 2 parents in 0 nuclear families
10 affected offspring trios
0 phenotypically discordant parent pairs found
Converting data to Individual-major format
Writing all Mendel errors to [ tinywga.mendel ]
Writing per-offspring Mendel summary to [ tinywga.imendel ]
Writing per-family Mendel summary to [ tinywga.fmendel ]
Writing per-locus Mendel summary to [ tinywga.lmendel ]
0 Mendel errors detected in total
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--out tinywga
--bfile /opt/galaxy/test-data/tinywga
--mind 1.0
--geno 1.0
--maf 0.0
--check-sex
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Converting data to Individual-major format
Writing X-chromosome sex check results to [ tinywga.sexcheck ]
Analysis finished: Wed May 19 15:15:38 2010
## Rgenetics: http://rgenetics.org Galaxy Tools rgQC.py Plink pruneLD runner
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile /opt/galaxy/test-data/tinywga
--indep-pairwise 40 20 0.5
--out tinywga
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 25 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.995
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 25 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Performing LD-based pruning...
Writing pruned-in SNPs to [ tinywga.prune.in ]
Writing pruned-out SNPs to [ tinywga.prune.out ]
Scanning from chromosome 22 to 22
Scan region on chromosome 22 from [ rs2283802 ] to [ rs4822375 ]
For chromosome 22, 11 SNPs pruned out, 14 remaining
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ ldp_tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile /opt/galaxy/test-data/tinywga
--extract tinywga.prune.in
--make-bed
--out ldp_tinywga
Reading map (extended format) from [ /opt/galaxy/test-data/tinywga.bim ]
25 markers to be included from [ /opt/galaxy/test-data/tinywga.bim ]
Reading pedigree information from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals read from [ /opt/galaxy/test-data/tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ /opt/galaxy/test-data/tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Reading list of SNPs to extract [ tinywga.prune.in ] ... 14 read
Before frequency and genotyping pruning, there are 14 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.991071
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 14 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
Writing pedigree information to [ ldp_tinywga.fam ]
Writing map (extended format) information to [ ldp_tinywga.bim ]
Writing genotype bitfile to [ ldp_tinywga.bed ]
Using (default) SNP-major mode
Analysis finished: Wed May 19 15:15:38 2010
@----------------------------------------------------------@
| PLINK! | v1.06 | 24/Apr/2009 |
|----------------------------------------------------------|
| (C) 2009 Shaun Purcell, GNU General Public License, v2 |
|----------------------------------------------------------|
| For documentation, citation & bug-report instructions: |
| http://pngu.mgh.harvard.edu/purcell/plink/ |
@----------------------------------------------------------@
Skipping web check... [ --noweb ]
Writing this text to log file [ tinywga.log ]
Analysis started: Wed May 19 15:15:38 2010
Options in effect:
--noweb
--bfile ldp_tinywga
--het
--out tinywga
Reading map (extended format) from [ ldp_tinywga.bim ]
14 markers to be included from [ ldp_tinywga.bim ]
Reading pedigree information from [ ldp_tinywga.fam ]
40 individuals read from [ ldp_tinywga.fam ]
40 individuals with nonmissing phenotypes
Assuming a disease phenotype (1=unaff, 2=aff, 0=miss)
Missing phenotype value is also -9
10 cases, 30 controls and 0 missing
21 males, 19 females, and 0 of unspecified sex
Reading genotype bitfile from [ ldp_tinywga.bed ]
Detected that binary PED file is v1.00 SNP-major mode
Before frequency and genotyping pruning, there are 14 SNPs
27 founders and 13 non-founders found
Total genotyping rate in remaining individuals is 0.991071
0 SNPs failed missingness test ( GENO > 1 )
0 SNPs failed frequency test ( MAF < 0 )
After frequency and genotyping pruning, there are 14 SNPs
After filtering, 10 cases, 30 controls and 0 missing
After filtering, 21 males, 19 females, and 0 of unspecified sex
**Warning** this analysis typically requires whole-genome level data
to give accurate results
Converting data to Individual-major format
Writing individual heterozygosity information to [ tinywga.het ]
Analysis finished: Wed May 19 15:15:38 2010